Background
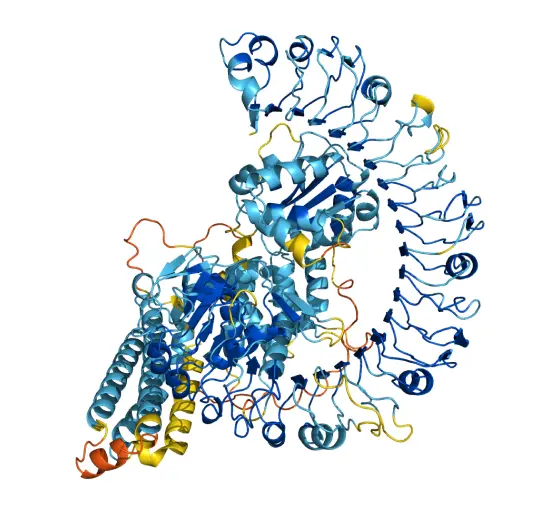
AlphaFold is an AI system developed by Google DeepMind that predicts a protein’s 3D structure from its amino acid sequence. It regularly achieves accuracy competitive with experiment.
Google DeepMind and EMBL’s European Bioinformatics Institute (EMBL-EBI) have partnered to create AlphaFold DB to make these predictions freely available to the scientific community. The latest database release contains over 200 million entries, providing broad coverage of UniProt (the standard repository of protein sequences and annotations). We provide individual downloads for the human proteome and for the proteomes of 47 other key organisms important in research and global health. We also provide a download for the manually curated subset of UniProt (Swiss-Prot).

Q8I3H7: May protect the malaria parasite against attack by the immune system. Mean pLDDT 85.57.
In CASP14, AlphaFold was the top-ranked protein structure prediction method by a large margin, producing predictions with high accuracy. While the system still has some limitations, the CASP results suggest AlphaFold has immediate potential to help us understand the structure of proteins and advance biological research.
Let us know how the AlphaFold Protein Structure Database has been useful in your research, or if you have questions not answered in the FAQs, at alphafold@deepmind.com.
If your use case isn't covered by the database, you can generate your own AlphaFold predictions using this open source code, which also supports multimer prediction.
What’s new?
Custom annotations - November 2025
We introduce a new functionality that enables users to integrate and visualise custom sequence annotations. This is made possible through the integration a protein feature web visualisation component.
The new feature, located on the "Annotations" tab, allows researchers to input single-residue annotations and region annotations. Custom annotations are visible on the 2D and 3D tracks, and alongside the predicted Local Distance Difference Test (pLDDT) score track.
What’s next?
We plan to continue updating the database with structures for newly discovered protein sequences, and to improve features and functionality in response to user feedback. Please follow Google DeepMind's and EMBL-EBI’s social channels for updates.
Licence and attributions
Data is available for academic and commercial use, under a CC-BY-4.0 licence.
EMBL-EBI expects attribution (e.g., in publications, services, or products) for any of its online services, databases, or software in accordance with good scientific practice.
If you use this resource, please cite the following papers:
If you use data from AlphaMissense in your work, please cite the following paper:
AlphaFold Data Provided by GDM:
AlphaFold Data Copyright (2022) DeepMind Technologies Limited.
AlphaMissense Copyright (2023) DeepMind Technologies Limited.
EMBL-EBI training
Recorded webinar
Accessing and interpreting predicted protein structures from AlphaFold database
Online tutorial